|
|
 |
|
| Biodynamics Laboratory Inc |
| Biodynamics Laboratory Inc.는 날마다 새로운 변혁이 일어나는 생명공학 분야의 연구자들을 위해 분자생물학 및 생화학 분야의 연구용 제품을 개발하고 있습니다. Biodynamics Laboratory Inc.는 genomics, proteomics, RNA research 등 생명과학 모든 분야에서 새로운 유전자, RNA의 새로운 역할, 단백질의 새로운 기능, 세포의 새로운 작용을 연구하는 연구자들에게 혁신적이고, 유용하며, 다루기 쉬운 바이오 연구의 도구를 제공합니다. |
| BioDynamics Laboratory Inc. 제품군 |
 DNA Size Marker, DNA Loading Dye, RNA Size Marker,Protein Size Marker DNA Size Marker, DNA Loading Dye, RNA Size Marker,Protein Size Marker |
 Easy to excise DNA bands from agarose gel without damage of DNA by UV Easy to excise DNA bands from agarose gel without damage of DNA by UV |
 Alkaline Phosphatase from psychrophilic bacterium Alkaline Phosphatase from psychrophilic bacterium |
 Tools for Molecular Biology & Life Science Tools for Molecular Biology & Life Science |
|
| Other reagents for molecular biology |
| DR110 |
DEPC-treated Water, 1 ml Χ 5 |
| DR115 |
DEPC-treated Water, 50 ml Χ 2 |
| DR117 |
DEPC-treated Water, 500 ml |
| DR120 |
RNase-free Water, non DEPC, 1 ml Χ 5 |
| DR125 |
RNase-free Water, non DEPC, 50 ml Χ 2 |
| DR127 |
RNase-free Water, non DEPC, 500 ml |
| F012 |
DNA, Salmon Sperm, Sonicated |
| F008 |
Anti-6 × Histidine Antibody |
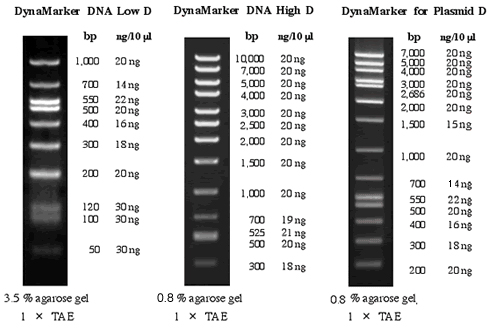
- 편리한 실험
- 연구자들이 사용하기 편리하도록 시약 내에 tracking dye가 포함되어 있습니다.
- Gel 상에서 DNA양의 간편한 측정
- DynaMarker® DNA Low D, DynaMarker® DNA High D, DynaMarker® for Plasmid D는
정확한 양의 DNA fragment들로 구성되어 있습니다.
- 높은 시인성
- UV 조명하에서 DNA의 size를 쉽게 구별하도록 하였습니다.
DynaMarker® Prestain Marker for Small RNA 시리즈는 electrophoresis와 blotting에서 작은 크기의 RNA를 측정하기 적합한 pre-stained molecular weight marker 입니다.
 |
 |
| Electrophoresis profile of DynaMarker® Prestain
Marker for Small RNA Plus (5 μl) on 10 % acrylamide containing
7.5 M urea gel with 1× TBE buffer as running buffer. |
Electrophoresis profile of DynaMarker® Prestain
Marker for Small RNA (5 μl) on 10 % acrylamide containing
7.5 M urea gel with 1× TBE buffer as running buffer. |
DynaMarker® Prestain Marker for Small RNA시리즈는 BioDynamics Laboratory Inc.의 고유 제품으로서 colored single-strand nucleic acid들로 구성되어 있습니다. DynaMarker® Prestain Marker for Small RNA시리즈의 각 band는 20, 30, 40, 50, 75, 100 base의 non-stained RNA와 일치합니다 (95% 정확도)
- 각 band는 20, 30, 40, 50, 75, 100 base의 non-stained RNA와 95% 정확도로 일치합니다 (figure 1)
- Prestained marker는 electrophoresis와 blotting 실험시 매우 유용합니다. (figure 2).
- 시인성이 우수한 파란색과 빨간색의 dual color indicator로 구성되어 있습니다.
- DynaMarker® Prestain Marker for Small RNA와 Small RNA Plus는 heat treatment나 denaturing agent가 필요치 않아 사용이 간편합니다.
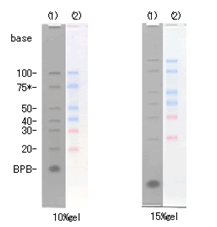 |
Figure 1
Electrophoresis profile of DynaMarker® Small RNA II + 75 base RNA* (1) and DynaMarker® Prestain Marker for Small RNA Plus (2) on 10 % and 15 % acrylamide containing 7.5 M urea gel / 1× TBE.
* 75 base RNA is from a newly synthesized RNA. A 75 base RNA is not included in DynaMarker® Small RNA II. |
 |
Figure 2
Left: Electrophoresis profile of DynaMaekr® Prestain Marker for Small RNA Plus (1) and RNA sample (2) on 10 % acrylamide containing 7.5 M urea gel / 1× TBE. Right: Blotting of (1) and (2) onto the nylon membrane. |
DynaMarker® Prestain Marker for RNA High는 electrophoresis와 blotting에서 큰 크기의 RNA를 측정하기 적합한 pre-stained molecular weight marker 입니다.
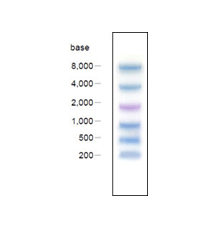 |
DynaMarker® Prestain Marker for RNA High는 RNA연구에 매우 유용한 도구입니다. 당 marker는 200, 500, 1,000, 2,000, 4,000, 8,000 base의 RNA에 대응하는 6개의 colored nucleic acid로 구성되어 있으며, nucleic acid chain으로 구성된 각 colored band는 band의 sharpness와 accuracy면에서 Bromophenol blue나 Xylenecyanol같은 dye와 다른 nucleic acid 고유의 특성을 보입니다. DynaMarker® Prestain Marker for RNA High는 electrophoresis와 blotting 연구에 적합하도록 제작되었습니다. |
- 90 %이상의 정확도. (figure 2)
- denaturing agarose gel electrophoresis와 nylon membrane blotting에 적합합니다(figure 3)
- 시인성이 우수한 파란색과 보라색의 dual color indicator로 구성되어 있습니다.
- DynaMarker® Prestain Marker for RNA High는 heat treatment나 denaturing agent가 필요치 않아 사용이 간편합니다.
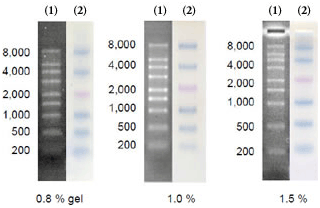 |
Figure 2
Electrophoresis profile of DynaMarker® RNA High(1) and DynaMarker® Prestain Marker for RNA High(2) on 0.8 %, 1.0 % and 1.5 % agarose - 2.2 M formaldehyde gel / 1x MOPS buffer as running buffer. |
 |
Figure 3
Left: Electrophoresis profile of DynaMarker® Prestain Marker for RNA High (1)and RNA sample (2) on 0.8 % agarose - 2.2 M formaldehyde gel / 1x MOPS buffer as running buffer. Right: Blotting of (1) and (2) onto nylon membrane. |
DynaMarker® RNA Low는 Denaturing PAGE에서 single-stranded RNA의 크기결정에 적합한 제품입니다.
- 20, 50, 100, 200, 300, 400, 500 base의 RNA size로 구성되어 있습니다.
- Maker의 각 RNA는 약 0.1 μg/μl의 동일한 양으로 구성되어 있어 Gel 상에서 RNA 시료의 대략적인 정량이 가능합니다
- siRNA의 분석을 위해 20-base RNA를 적용하였습니다
|

DynaMarker® RNA Low II
Electrophoresis profile of DynaMarker® RNA Low II (0.7μg) on 5% of acrylamide, 8 M urea gel with 1×TBE buffer as running buffer
|
Loading이 간편한 DynaMarker® RNA Low II는 Denaturing PAGE에 적합한 제품입니다..
- 20, 50, 100, 200, 300, 400, 500 base의 RNA size로 구성된 Ready-to-load RNA marker
- Maker의 각 RNA는 약 0.1μg/5μl로 구성되어 있습니다.(well당 적정 loading 양은 2.5-5 μl입니다.)
- RNA Loading Buffer PA는 denaturing PAGE에서 쉬운 sample preparation을 제공합니다
|

DynaMarker® RNA Low II
Electrophoresis profile of DynaMarker® RNA Low II (0.7μg) on 5% of acrylamide, 8 M urea gel with 1×TBE buffer as running buffer
|
DynaMarker® RNA Low는 Denaturing PAGE에서 single-stranded RNA의 크기결정에 적합한 제품입니다.
- 200, 500, 1000, 1500, 2000, 3000, 4000, 5000, 8000 base 의 RNA size로 구성되어 있습니다.
- Maker의 각 RNA는 약 0.1 μg/μl의 동일한 양으로 구성되어 있어 Gel 상에서 RNA 시료의 대략적인 정량이 가능합니다
|

DynaMarker® RNA High
Electrophoresis profile of DynaMarker® RNA High (0.9μg) on formaldehyde-agarose (1%) gel
|

DynaMarker® RNA Easy Measurement N은 denaturing 뿐만 아니라 non-denaturing agarose gel electrophorsis에서의 RNA 크기결정에 적합한 제품입니다.
- DynaMarker® RNA High AGN은 200, 500, 1000, 1500, 2000, 3000, 4000, 5000, 8000 base의 RNA size로 구성되어 있습니다
- Maker의 각 RNA는 약 0.1 μg/μl의 동일한 양으로 구성되어 있어 Gel 상에서 RNA 시료의 대략적인 정량이 가능합니다
- Agarose gel electrophoresis에서 간편한 RNA preperation을 위해 RNA Loading Buffer AG가 제공됩니다. RNA Loading Buffer AG는 denaturing 뿐만 아니라 non-denaturing agarose gel(1X TAE, 0.5X TBE)에서 RNA electrophorsis가 가능합니다.
|

DynaMarker®
RNA Easy Measurment N
Electrophoresis profile of DynaMarker® RNA High
AGN on formaldehyde-agarose (1%) gel. Left lane : 0.45 μg of DynaMarker® RNA High AGN Right lane : 0.4 μg of Human Total RNA.
|
- Profile of electrophoresis

Denaturing agarose gel |
Non-denaturing agarose gel |
DynaMarker® RNA High AGN (0.45 μg/ well), Human Total RNA (0.4 μg/ well) were electrophoresed on denaturing agarose gel (left) or non-denaturing agarose gel (right). DynaMarker® RNA High AGN or Human Total RNA was mixed with formaldehyde-added RNA Loading Buffer AG+ and treated as above.
DNA fragments for DynaMarker® RNA High Probe는 DynaMarker® RNA High와 DynaMarker® RNA High AGN (in DynaMarker® RNA Easy Measurement N)의 hybridization 실험의 검출에 적합한 제품입니다.
- DynaMarker® RNA High와 DynaMarker® RNA High AGN (in DynaMarker® RNA Easy Measurement N)의 모든 RNA band를 검출합니다.
- Probe의 isotopic 및 non-isotopic labeling에 모두 적합합니다.
- 170 - 200 bp의 of 5' phosphorylated dsDNA fragments (5'-protruding ends)로 구성되어 있습니다.
Profile of hybridization
 |
Northen hybridization with DNA fragments for DynaMarker® RNA High Probe
DynaMarker® RNA High was electrophoresed in formaldehyde-agarose (1%) gel and transferred onto nylon membrane. DNA fragments for DynaMarker® RNA High Probe was labeled by non-isotopic method and hybridized to the nylon membrane. After washing the blot, it was reacted with chemiluminescence substrate. Signal was exposed to a high speed film. |
Non-denaturing PAGE에서 double-stranded RNA의 크기결정에 적합한 제품입니다.
- 10, 20, 30, 50, 100, 200, 300, 400, 500, 1000 bp의 dsRNA size로 구성되어 있습니다.
- 20 bp dsRNA의 농도는 25 ng/μl로 적정되어 있습니다. 2μl의 DynaMarker® dsRNA에는 siRNA 분석에 적합한 20 bp dsRNA 50 ng이 포함되어 있습니다.
Non-denaturing PAGE 실험을 위한 Ready-to-load DynaMarker® dsRNA
- Ready-to-load dsRNA marker는 10, 20, 30, 50, 100, 200, 300, 400, 500, 1000 bp의 dsRNA size로 구성되어 있습니다.
- 20 bp dsRNA의 농도는 10 ng/μl로 적정되어 있습니다.
5μl의 DynaMarker® dsRNA에는 siRNA 분석에 적합한 20 bp dsRNA 50 ng이 포함되어 있습니다.
- 간편한 dsRNA sample preparation을 위해 6 × dsRNA Loading Buffer가 제공됩니다.
| DynaMarker® Small RNA II는 denaturing PAGE에서 single-stranded RNA (ssRNA)의 작은 크기를 결정하기 적합한 molecular weight marker입니다.
- 20, 30, 40, 50 and 100 base의 ssRNA로 구성되어 있습니다.
- siRNA와 miRNA 분석에 유용합니다.
- Denaturing PAGE에서의 높은 해상도를 위해 각 band는 높은 순도로 정제되어 있습니다
|

DynaMarker® Small RNA II
Electrophoresis profile of DynaMarker® Small RNA II (1μl) on 12.5% of acrylamide, 7.5 M urea gel with 1×TBE buffer as running buffer |
| DynaMarker® Small RNA II Easy Load 는 denaturing PAGE에서 single-stranded RNA (ssRNA)의 작은 크기를 결정하기 적합한 molecular weight marker로서 loading buffer가 포함되어 있는 ready-to-use type의 제품입니다.
- 20, 30, 40, 50 and 100 base의 ssRNA로 구성되어 있습니다.
- siRNA와 miRNA 분석에 유용합니다.
- Denaturing PAGE에서의 높은 해상도를 위해 각 band는 높은 순도로 정제되어 있습니다.
- RNA Loading Buffer PA는 denaturing PAGE에서 쉬운 sample preparation을 제공합니다.
|

DynaMarker® Small RNA II Easy Load
Electrophoresis profile of DynaMarker® Small RNA II Easy Load (5 μl) on 12.5% of acrylamide, 7.5 M urea gel with 1× TBE buffer as running buffe |
DynaMarker® Protein Eco는 SDS-PAGE에서 단백질 분자량 결정에 유용한 제품입니다
- 분자량이 잘 알려진 표준 단백질들로 구성되어 있습니다.
- SDS-PAGE에서 간편한 sample preparation를 위해 Protein Loading Dye가 제공됩니다.
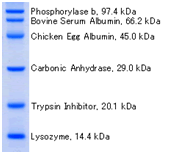
DynaMarker® Protein Eco (2.5 μl) was runon SDS-PAGE (12%) |
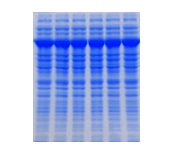
Recobinant E. coli is lysed with Protein Loading Dye and run on SDSPAGE (10%). Expressed protein is seen as thick bands. |
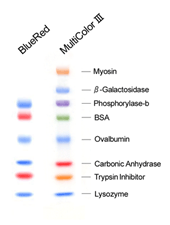 DynaMarker® ProteinBlueRed or MultiColor III
DynaMarker® ProteinBlueRed or MultiColor III is run on a 5-20% acrylamide gradient gel according to the method of Laemmli.
DynaMarker® Protein BlueRed와 Protein MultiColor III은 staining 과정 없이 SDS-PAGE에서 단백질 분리과정의 진행과 western blotting에서 blotting 효율을 확인하기 위한 prestained-marker입니다.
DynaMarker® Protein BlueRed는 파란색과 빨간색의 dual-color size marker로 구성되어 있으며, DynaMarker® Protein MultiColor III는 보라색, 파란색, 초록색, 빨간색, 주황색 5가지 색상의 size marker로 구성되어 있습니다.
- Multiple color로 인해 즉각적인 band의 인식이 가능합니다
- 각각의 marker들은 밝고 선명한 band를 나타냅니다.
- 각각의 marker들은 고순도로 정제된 단백질에 동일한 정도의 dye 염색을 거쳐 제조됩니다. Coomassie-dye staining후에도 선명하고 균일한 band를 관찰할 수 있습니다.
- 당 제품은 staining 없이 western blotting의 진행을 monitor하는데 유용합니다

Before staining |
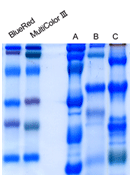
After Coomassie Staining |
Comparison markers of prestained DynaMarker® Protein series and other commercial multi-color prestained marker (non-recombinant).
Each marker is run on a 12.5% polyacrylamide gel according to the standard method. Markers of prestained DynaMarker® Protein series cover adequate molecular weight range and offer bright-color and sharp bands. Because highly purified proteins in the marker were covalently and stoichiometrically bonded with high quality dye and protein is adjusted to approximate equal amount, after coomassie-dye staining sharp and uniform bands appear without extra bands.
DynaMarker® Protein Recombinant은 10 kDa에서 150 kDa range의정확한분자량의 recombinant protein 9개로구성되어있습니다
- 10 kDa ~ 150 kDa range..
- 9개의 재조합 단백질로 구성
- 10, 20 kDa standard에 의한 쉬운 size 계산
- Glycosylation없는 선명한 band.
- Gel loading buffer와 혼합되어 있어, loading이 용이합니다.
- 두 개의 진한 band(20 kDa and 80 kDa)가 있어 size 인식이 용이합니다.
|

DynaMarker® Protein Recombinant 5 μl
5-20 % Gradient Gel |
DynaMarker® Protein Recombinant MultiColor는 10 kDa에서 150 kDa range의 pre-stained recombinant protein로구성되어있습니다. 각각의 marker는파란색, 보라색, 초록색, 빨간색, 주황색으로염색되어있어 protein band의인식이용이한제품입니다.
- 15 kDa ~ 150 kDa range.
- 선명한 다섯가지 색상(파란색, 보라색, 초록색, 빨간색, 주황색)의 표준단백질
- 색상으로 인해 각각의 band 구별이 용이
- 10, 20 kDa standard에 의한 쉬운 size 계산
- 각각의 band 분자량의 정확도는 95% 이상입니다
- Gel loading buffer와 혼합되어 있어, loading이 용이합니다.
|
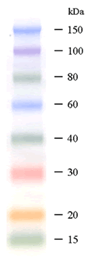
DynaMarker® Protein Recombinant 5 μl
5-20 % Gradient Gel |

UV에의한 DNA 손상없이 agarose gel로부터 DNA band를추출합니다.
- 실험대에서 직접 DNA를 추출합니다.
- 유해한 Ethidium bromide없이 DNA band 확인이 가능합니다.
- UV를 사용하지 않아, DNA의 손상이 없습니다.
- 시판중인 모든 DNA purification kit에 적용할 수 있습니다.
- 고감도 DNA 검출방법입니다.
|

In the picture, 2 kbp of DNA fragment is used
|
Ligation and Transformation using DNA excised withGel Indicator™
 |
About a 500 bp of DNA fragment obtained by enzyme digestion was separated on agarose gel electrophoresis and were excised, with Gel Indicator™ (see figure, 0 min) or on UV illuminator after staining of ethidiumbromide. During UV exposure (see figure, 3, 5, 10 min) for excision DNA bands, a gel strip was turn over occasionally. DNA was extracted from an excised gel strip by commercially available spin kit. The concentration of each obtained DNA was measured and the same amount of DNA was added to a plasmid vector. After ligation of these DNA mixture with T4 ligase, they were used for E. coli transformation. E. coli cells were inoculated on LB agar plates containing antibiotic and incubated at 37°C, overnight. The figure shows the number of white colonies on these LB agar plates.
In the experiment used with Gel Indicator™, higher efficiency of transformation is shown |
 UV 손상없는 DNA 추출과고감도 DNA 검출
UV 노출에 의한 DNA의 손상은 transcription, PCR, cloning 등의 이후 실험에 예상치 못한 결과를 초래합니다(1, 2). Gel Indicator™ DX은 고감도의 검출한계(> 20ng)와 UV에 의한 손상 없는 DNA 추출방법을 제공합니다
1. Cariello NF, Keohavong P, Sanderson BJ, Thilly WG., Nuc. Acids. Res., 16 (1988) 4157.
2. Hartman P.S., Biotechniques, 11 (1991) 747-748.
- 고감도 DNA 검출: > 20 ng
- UV를 사용하지 않아, DNA의 손상이 없습니다.
- UV 없이 DNA의 확인이 가능합니다
Higher Cloning Efficiency with Gel Indicator™ DX
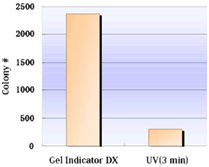
Fig.1 Transformation Efficiency
|
Transformation Efficiency
DNA fragment (2,000 bp) was separated on agarose gel electrophoresis and were excised, with Gel Indicator™ DX (see left bar of the figure) or on UV illuminator after staining of ethidiumbromide(UV exposure for 3 min, right bar). DNA was extracted from an excised gel strip. The concentration of each obtained DNA was measured and the same amount of DNA was used for ligation. E. coli cells were transformed with these ligated DNAs. |
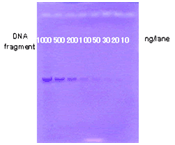
Higher Sensitivity of Gel Indicator™ DX

Fig.2 Detection of DNA by Protocol II
|
Detection of DNA by Protocol II
Serially diluted DNAs were loaded to agarose gel (0.8 %) containing Gel Indicator Solution DX. After Electrophoresis, the gel was colorized with GI Coloring Solution for high sensitive detection. |
Gel Indicator™와함께사용되어 DNA 추출과정제를동시에제공하는제품입니다.
Gel Indicator™ DNA Extraction Kit을 사용하면 UV에 의한 손상 없이 agarose gel로부터 DNA를 추출 정제할 수 있습니다. UV 손상 없이 추출된 DNA는 cloning, enzyme digestion, sequencing, PCR등의 응용에 적합하게 사용됩니다. Electrophoresis후 즉시 추출하는 rapid method와 50ng까지 검출할 수 있는 high-sensitive DNA detection method의 두 가지 protocol을 제공합니다.
- UV 손상없는 DNA가 정제됩니다.
Ethidium bromide 없이 DNA band를 확인합니다.
DNA가 고효율로 cloning됩니다.
- Agarose gel로부터 추출시 1 × TAE buffer 및 0.5 × TBE buffer의 사용이 가능합니다.
- 정제된 DNA는 cloning, enzyme digestion, sequencing, PCR등의 응용에 적합하게 사용됩니다.
- DNA의 수율은 70~80%입니다.
- DNA cloning을 위해 당 제품은 circular DNA보다 linear DNA의 정제에 적합하도록 제작되었습니다. (circular DNA의 효율은 10%이하입니다.)
|
 |

|
|
| Fig. 1. Electrophoresis by protocol I |
Fig. 2. Detection of DNA by Protocol II.
Serially diluted DNA fragment (2000 bp) was
loaded to wells from right to left lanes.
Agarose Electrophoresis (0.8 %), 1 × TAE |
| 1. |
Weight a gel slice and transfer it to a tube. |
| 2. |
Add 3 volume of Gel Dissolving Buffer. |
| 3. |
Incubate a tube containing gel and Gel Dissolving Buffer at 50-55°C. |
| 4. |
After dissolving gel completely (5 min or more),add one gel volume of isopropanol to the dissolved gel and mix well. |
| 5. |
Load the dissolved gel to a spin column. Centrifuge, 1 min at 7,000 -10,000 × g. Discard the filtrate. |
| 6. |
Wash with 700 μl of Wash Buffer (+EtOH) |
| 7. |
Centrifuge, 1 min at 7,000 -10,000 × g. Discard the filtrate. |
| 8. |
Centrifuge again, 1 min at 7,000 -10,000 × g. |
| 9. |
Transfer the Spin Column to a new tube. Load 50 μl of Elution Buffer onto the Spin Column to elute. Centrifuge, 1 min at 7,000 -10,000 × g.
|
Ligation and Transformation by DNA purified with Gel Indicator™ DNA Extraction Kit
| DNA fragment, about a 2 kbp, resulted from enzyme digestion was separated on agarose gel electrophoresis. DNA were excised by the method of Gel Indicator™ DNA Extraction Kit. One of DNA in gel was extracted and purified by the method of Gel Indicator™ DNA Extraction Kit. (see figure, 0 min). Other DNA in gel was exposed to the ultraviolet ray (see figure, 1, 3, 5, 10 min) after staining of ethidiumbromide and purified with Gel Indicator™ DNA Extraction Kit. During UV exposure ( see figure, 1, 3, 5, 10 min) for excision DNA bands, a gel slice was turn over occasionally. The concentration of each obtained DNA was measured and the same amount of DNA was added to a plasmid vector. |
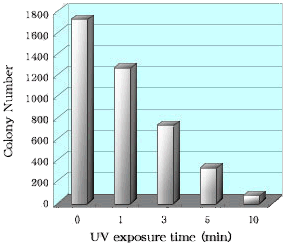 |
| After ligation of these DNA mixture with T4 ligase, they were used for E. coli transformation. E. coli cells were inoculated on LB agar plates containing antibiotic, X-gal and IPTG. at 37°C, overnight. The figure shows the number of white colonies on these LB agar plates. This method of Gel Indicator™ DNA Extraction Kit shows much higher efficiency of transformation than the usual method of ultraviolet-ray exposure. |
 Gel Indicator™RNA Staining Solution은 RNA의추출을위해 PAGE gel의 RNA를염색하는제품으로서사용이간편한 ready-to-use type solution입니다. RNA의검출한계는 50ng으로서 UV shadowing에비해 5배가랑우수한감도를보이며, 20 mer 가량의작은 RNA도검출할수있는제품입니다
- 간편한 ready-to-use solution (staining time; 20-30 min)
- Transilluminator 없이, RNA band를 육안으로 확인합니다.
- RNA의검출한계는 50ng으로서 UV shadowing에비해 5배가랑우수한감도를보입니다.
- 염색된 RNA는 gel로부터 crush and soak method와 ethanol precipitation과정을 통해 추출합니다. 추출한 RNA는 RT-PCR, enzyme reaction, labeling reaction등이 바로 사용할 수 있습니다.
Sensivity of Gel Indicator™ RNA Staining Solution

Fig.1 |
Fig.1 Staining with Gel Indicator™ RNA Staining Solution
Serially diluted small RNAs (21 base) were subjected to denaturing polyacrylamide gel electrophoresis (12.5 % of polyacrylamide gel containing 7.5 M of Urea, 1× TBE as running buffer) After Electrophoresis, the gel was stained with Gel Indicator™ RNA Staining Solution. It detected 50 ng of 21-base RNA.
|
Recovery of RNA with Gel Indicator™ RNA Staining Solution

Fig.2 |
Fig. 2 Recovery of RNA with Gel Indicator™ RNA staining Solution
RNA (100 base) prepared by in vitro transcription was subjected to denaturing-polyacrylamide gel electrophoresis. The RNA was excised and extraction by a crush and soak method after staining with Gel Indicator™ RNA Staining Solution. Obtained RNAs was analyzed by denaturing-polyacrylamide gel electrophoresis (5 % of polyacrylamide gel containing 8 M of Urea, 1× TBE as running buffer). Recovered RNA from gel with using Gel Indicator™ RNA staining Solution showed that the RNA was of high integrity.
Lane 1: RNA prepared by in vitro transcription
Lane 2: Gel-purified RNA
|

Psychrophilic strain Shewanella sp. SIB1 로부터얻은 Alkaline phosphatase (PAP)는 BAP와 CIAP의 장점을 모두 갖고 있습니다.
 |
- Psychrophilic strain Shewanella sp. SIB1 로부터얻은 Alkaline phosphatase (PAP)는 SAP나 CIAP처럼 heat inactivation시킬 수 있습니다.
반면에, BAP는 phenol extraction으로도 inactivation 시키기 어렵습니다.
- 60°C에서의 enzyme activity는 37°C에 비해 4배 가량 높기 때문에, protruding 3'-end 뿐만 아니라 blunt end or recessed 5'-end의 phosphate도 효과적으로 제거할 수 있습니다.
(반면에, SAP and CIAP는 60°C에서 inactivation 됩니다.)
|
PAP의장점
BIP는 inactivation시키기 어려운 반면에, PAP는 heat inactivation이 가능합니다.
아래 그림에서 보여지듯이, transformation시 PAP의 처리는 BAP에 비해 plate의 많은 white colony를 보입니다. BAP는 inactivation treatment후에도 상당량이 남아있기 때문에 active BAP는 ligation reaction 시 insert DNA의 phosphate를 제거하여 ligation 효율을 떨어트립니다. 아래 실험은 ligation 전에 PAP를 효과적으로 inactivation시키는 것을 보여줍니다. BAP 처리 후에 충분한 양의 white colony를 얻기 위해서는 ligation 시 high ratio의 insert vector를 처리하거나 2회 이상의 phenol extraction을 거쳐야 합니다.
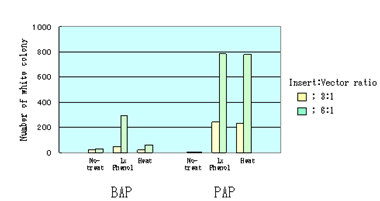
One μg of the EcoRI cleaved pBluescript SK (+) vector was dephosphorylated by 0.5 unit of BAP or 5 unit PAP at 37°C. After dephosphorylation, the reaction mixtures were treated as follow,
| 1. |
No-treatment: The reaction mixtures were directly used for ligation reaction. |
| 2. |
Phenol: Equal volume of phenol was added to the reaction mixture and it was vortexed for 30 seconds. Then the mixture was extracted by ether, and precipitated by ethanol. The precipitate was dried up and dissolved by dH2O. It was used for ligation reaction. |
| 3. |
Heat: The reaction mixtures were just heat at 95°C for 5 min for PAP or at 100°C for 5 min for BAP. These were directly used for ligation reaction. |
After above treatments, EcoRI cleaved, dephosphorylated pBluescript SK (+) vector were ligated to a 1kb insert DNA fragment. Competent XL1-Blues were transformed with the ligation products. Number of white colony was shown in figure.
Thermotoga maritima의 RNase는 pre-tRNA로부터 3' trailer를제거하여 tRNA의 3'-terminal CCA를생성합니다.
Minagawa et al., Journal of Biological Chemistry, 279 (2004) 15688

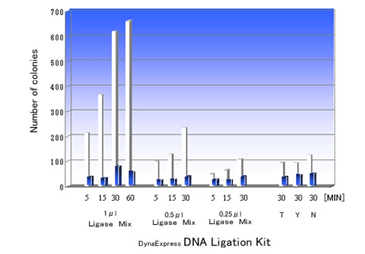
DNA Ligation Kit ver.2는 cohesive end 또는 blunt end DNA fragments 을 16°C~25°C에서 5-30분내에효과적으로 ligation시킵니다. Ligation reaction은 vector와 insert DNA의혼합물에 2 × Ligation Buffer 와 Ligase Mixture를넣는것으로간단히시작되며, ligation reaction mixture는 competent cell의 transformation에바로사용할수있습니다.
- 높은 ligation 효율.
- 간단한 과정.
- 짧은 반응 시간 (5-30 분).
- Ligation reaction후 transformation에 바로 사용 가능
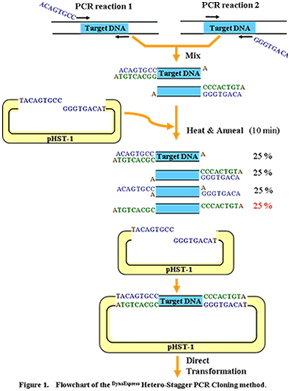
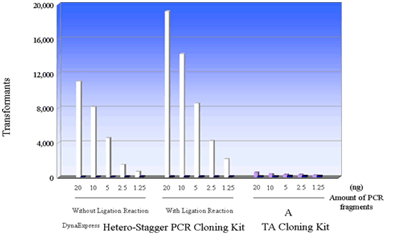
DynaExpress Hetero-Stagger PCR Cloning Kit는 PCR product의효과적이고빠른 cloning을가능하게합니다. 이새로운방법은 restriction enzyme, ligase, exonuclease, Urasil DNA glycosylase, Cre-loxP recombinase reactions과같은 enzymatic reaction을필요로하지않는대신 4개의 PCR primer를사용합니다. 2개의 PCR primer에는 non-proofreading 또는 proofreading thermostable DNA polymerase 여부에따라 5' end에 8~9개의 extra base가있으며, 다른 2개의 PCR primer에는 extra base가없습니다. 각기다른 extra terminal sequence를갖는 2개의 PCR product를 2번의 PCR reaction으로얻은후, 두 PCR product를 pHST vector와혼합하고 heat denaturation, annealing을거쳐 pHST vector의 single-strand extension과상보적인 9 base overhang을갖는 heteroduplex product를만듭니다. Vector와 insert의 complementary extension이충분히길어 ligation이필요치않으며, vector와 insert의 mixture는 competent cell의 transformation에바로사용될수있습니다.
- 높은 ligation 효율.
- 단순한 원리
- 간단한 과정 (mix, heat and anneal)
- Enzymatic reaction이 필요 없습니다.
- Annealing후 직접 transformation에 사용 가능합니다.
- Insert orientation의 선택이 자유롭습니다.
- Proofreading 또는 non-proofreading DNA polymerase에 모두 적용 가능합니다
- PCR product부터 plating까지 90분이면 충분합니다.
The effect on cloning using different amounts of PCR fragments (about 1kb) was tested regarding the DynaExpress Hetero-Stagger PCR Cloning Kit and a TA cloning Kit (Supplier A). The recommended protocol from each supplier was followed. After the reaction between PCR fragments and vector, competent cells were transformed with 1/3 amount of reaction mixture. Half of the amounts of the cells were plated onto the agar plate.

 |
최근약 18~28 nucleotide 길이의 non-coding RNA가많은관심을받고있으며 short interfering RNA (siRNA)와 microRNA (miRNA)가 non-coding RNA의주요 category입니다. miRNA는 eukaryote에널리분포하며 differentiation, development, viral infection등의 regulatory molecule로써작용합니다.
DynaExpress miRNA Cloning Kit High Efficient는 non-coding RNA의 cloning을 위한 제품입니다.
- Small RNA (18-28 nucleotide)의 cloning을 위한 ready-to-use reagent
- 3' linker 또는 5' linker와의 효과적인 ligation을 위한 specific ligase.
- Ligase 3'과 activated MI-A3’ Linker를 이용한 5’ - phosphorylated miRNA와 3’ linker의 ligation
- Isotope의 사용이 없어 안전합니다.
- Color guided size marker를 이용한 간편한 linker-ligated miRNA purification
- Step-by-step protocol
- Total RNA로부터 small RNA를 분리하기 위한 2 step과 small RNA cloning을 위한 5 reaction
|
|